Misactivation of Developmental Enhancers for the SOX2 Transcription Factor in Breast and Lung Cancer
This paper on @biorxivpreprint explores how misactivation of developmental enhancers for the SOX2 transcription factor leads to overexpression in breast and lung cancer. Learn about the promoter, potential enhancers, and more.

Michael Hoffman @michaelhoffman@mastodon.social
Computational genomicist, Princess Margaret Cancer Centre. Assoc Prof, Medical Biophysics & Computer Science, University of Toronto. COI https://t.co/CvupKNIjZr
-
1/Now on @biorxivpreprint: uncovering how misactivation of developmental enhancers for the SOX2 transcription factor leads to overexpression in breast and lung cancer. By Luis Abatti, P. Lado-Fernández, @vietlinh_huynh, @mcollado_CHUS, me, @mitchell_lab. https://t.co/7uzyMkEQYy
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023 -
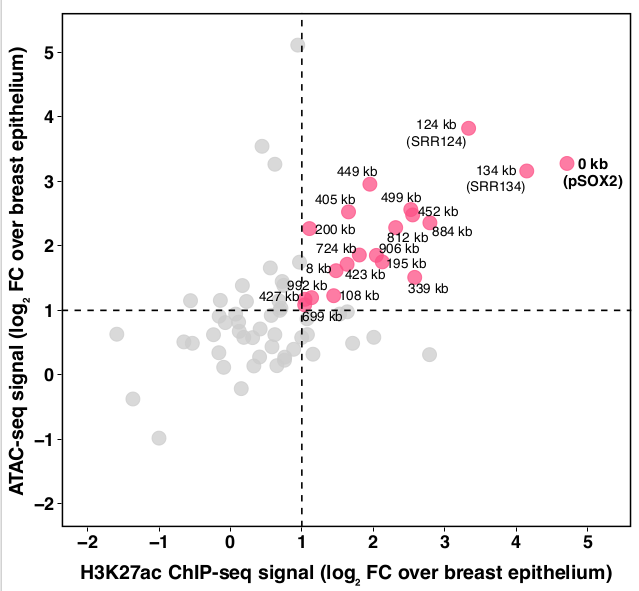
4/In MCF-7 luminal A breast cancer cells, 19 regions gained putative enhancer/promoter features (H3K27ac+open chromatin) compared to healthy breast epithelium.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023
pSOX2 is the promoter. Next highest are potential enhancers 124 and 134 kbp downstream, which we call SRR124 & SRR134. pic.twitter.com/gQ90i2uUw6 -
5/@ENCODE_NIH data show the presence of multiple (SRR124) and many (SRR134) transcription factors in MCF-7 cells, a key feature of active enhancers. pic.twitter.com/hHiM2lt2XN
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023 -
6/A similar story in other cell lines modeling lung squamous carcinoma, lung adenocarcinoma, and luminal A breast carcinoma: development of enhancer features at SRR124 and SRR134.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023
ChIA-PET data show how these features are linked in 3D genome organization to the SOX2 promoter. pic.twitter.com/mellS6ZyUN -
7/Also not much in the way of changes at previous described enhancer regions SRR1, SRR2, and hSCR. In summary, the SRR124–134 cluster seems to be key in SOX2 tumorigenic overexpression.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023 -
8/Enhancer constructs transfected into the same four cancer cell lines show the activation of this cluster, but no significant activation of the previously studied enhancers SRR1, SRR2, and hSCR. pic.twitter.com/wE5DmxL08m
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023 -
9/Heterozygous CRISPR/Cas9 deletion of this enhancer cluster in MCF-7 and PC-9 reduces SOX2 expression by 60%.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023
Homozygous deletion reduces SOX2 expression to nearly nothing. pic.twitter.com/wkedaHWesw -
10/Homozygous SRR124–134 enhancer cluster knockouts greatly reduced the ability of the cells to form colonies, suggesting that these cells require the SOX2 overexpression driven by this cluster for high tumor initiation capacity. pic.twitter.com/PpLljjK1N5
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023 -
11/Differential expression analysis of the homozygous deleted enhancer MCF-7 versus wild-type shows 529 genes with changes in expression, with the largest change for SOX2 itself. pic.twitter.com/uhvWeiC1e7
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 15, 2023 -
12/Homozygous deletion of the enhancer cluster led to changes in chromatin accessibility at 3,076 500-bp regions. pic.twitter.com/2TrpTEcltE
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
13/TOBIAS analysis showed 281 motifs with differential transcription factor footprints in ATAC-seq peaks, almost all of which were decreases in apparent binding, indicating how reduced SOX2 expression affects binding of multiple other transcription factors. pic.twitter.com/eoLp4c3kFX
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
14/In #TCGA cancer patient data, the SRR124–134 enhancer cluster is most accessible in:
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023
- lung squamous cell carcinoma (LUSC)
- lung adenocarcinoma (LUAD)
- breast carcinoma (BRCA)
- bladder carcinoma (BLCA)
- stomach adenocarcinoma (STAD)
_ uterine endometrial carcinoma (UCEC) pic.twitter.com/hOVeM2ykth -
16/The SRR124–134 enhancer cluster show increased accessibility across multiple TCGA tumors of these types, unlike other previously described enhancers. pic.twitter.com/GulwuGSNLi
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
17/In these cancer types, increased SRR124–134 accessibility associates with higher pSOX2 accessibility and higher SOX2 expression. pic.twitter.com/aSsWFFhwFf
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
18/In BRCA, LUAD, and LUSC, expression of 115 transcription factors significantly associate with SRR124 chromatin accessibility, and 90 transcription factors associate with SRR134 accessibility, including FOXA1 (positive correlation for both) and NFIB (negative for both). pic.twitter.com/nGn4s4qFUa
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
19/FOXA1 expression significantly and positively correlates with both SRR124 and SRR134 accessible chromatin. NFIB expression significantly and negatively correlates with accessible chromatin at both. pic.twitter.com/skO1glJV7W
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
20/Laboratory overexpression of FOXA1 led to increased SRR124 and SRR134 enhancer construct activity in four cell lines. Overexpression of NFIB led to decreased enhancer construct activity in MCF-7, T47D, and H520. pic.twitter.com/IAcLj6IMbE
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
21/Mutating a FOXA1 motif in SRR134 led to greatly reduced enhancer activity, indicating that it is crucial for sustaining SRR134 activity in these cancer contexts. NFIB seems more dispensible. pic.twitter.com/ZLg3I4OCNe
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
22/Since SOX2 is required for proper development of multiple tissues, we examining the accessibility of SOX2 enhancers across multiple tissues. This revealed that, beyond cancer, SRR124 and SRR134 are accessible in digestive and pulmonary tissue. pic.twitter.com/YeQVS4nYh1
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
23/We also found that the SRR124 core sequence is highly conserved in the mouse (we call the homolog mSRR96), as is SRR134 (mSRR102). pic.twitter.com/QXY7U9GePN
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
24/We generated a mouse knockout spanning the mSRR96–102 enhancer cluster, and crossed deletion heterzygotes. No homozygous offspring lived until weaning, indicating this enhancer cluster is critical for mouse survival. pic.twitter.com/yjhu8CN5uU
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
25/Examination of E18.5 embryos showed that deletion heterozygotes developed esophageal atresia/tracheoesophageal fistula (EA/TEF)—the esophagus and trachea failed to separate during development. The SOX2 protein was completely absent in this tissue. pic.twitter.com/B9eIRyKH6i
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
26/Wild-type and deletion heterozygotes had both high levels of SOX2 protein in the esophageal and tracheal tubes.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
27/In summary:
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023
- these particular SOX2 enhancers are essential during normal development
- their repurposing is essential in particular cancers
I'd argue that:
- deciphering how enhancer misactivation causes tumorigenesis is essential for reversing or stopping it -
28/The technology now available for genome editing and examining subsequent changes in gene regulatory behavior makes this possible like never before.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 16, 2023 -
29/MCF-7 RNA-seq and ATAC-seq data with and without the enhancer deletion are available on @NCBI GEO. https://t.co/xuMlPzJ6gJ
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
30/Other data are available in 29 supplementary tables, extensively documented with a data dictionary describing them. pic.twitter.com/5vSTcUFqW0
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
31/Thanks to @CIHR_IRSC, @InnovationCA, and @ONtrainandstudy for funding.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023
