Modeling Methyl-Sensitive Transcription Factor Motifs
Learn about a method to explicitly model how cytosine modifications on one side of a base pair or the other might affect transcription factor binding. Understand how to expand the alphabet and develop a model to understand the effects.

Michael Hoffman @michaelhoffman@mastodon.social
Computational genomicist, Princess Margaret Cancer Centre. Assoc Prof, Medical Biophysics & Computer Science, University of Toronto. COI https://t.co/CvupKNIjZr
-
1/On @biorxivpreprint: revised preprint from my lab: modeling methyl-sensitive transcription factor motifs with an expanded epigenetic alphabet. By @cobyviner @CharlesIshak @marcela_sjoberg @roxs88 @David_J_Adams @AnneF_S @decarvalho_lab @HainerLab et al! https://t.co/QHDJSlgDxm
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
3/While these modifications do not alter DNA base pairing, they do change DNA conformation, and therefore can and do affect how transcription factors bind at this position.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
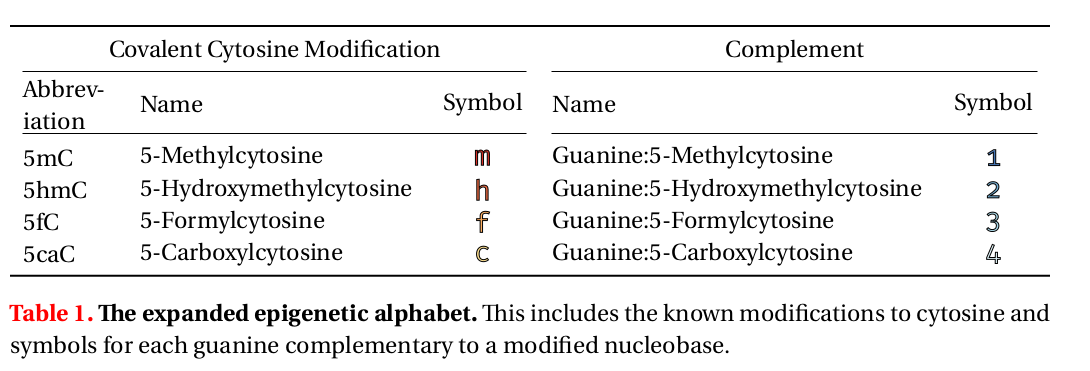
4/We developed a method to explicitly model how cytosine modifications on one side of a base pair or the other might affect transcription factor binding. To do this we simply expand the alphabet, adding m for methylcytosine, h for hydroxymethylcytosine, and so on. pic.twitter.com/fRXhflstkk
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
5/We have added the capability to have custom alphabets, and this custom alphabet to the MEME Suite. So anyone can use it already. https://t.co/H6WAT80GjU
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
6/Since epigenetic modification data comes from a population of cells, you often will not have 100% of reads at a position indicating the modification. So we have to use a threshold to decide whether a base should be called C or m.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
7/From @blueprint_eu bisulfite sequencing and oxidative bisulfite sequencing data, we called modifications at many thresholds. We examined how that affected enrichment of unmodified vs modified motifs in ChIP-seq peaks for transcription factors with known modification preferences
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
8/Specifically, we looked at
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023
- c-Myc, known to prefer unmethylated DNA
- ZFP57, known to prefer methylated DNA
- C/EBPβ, known to prefer methylated DNA
In each case, the difference in enrichment score observed reflected these known preferences, over every threshold examined. pic.twitter.com/WfuZQQKjDa -
9/Interestingly, with oxidative bisulfite sequencing data, our method also teased apart its previously reported, but less widely known ability to distinguish between 5mC and 5hmC. pic.twitter.com/UqfWtni9tM
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
10/Examining ChIP-seq data for 144 transcription factors, we found some that bind in both modified and unmodified contexts, but sometimes have rather different motifs when doing so. Expanding the alphabet allows us to identify previously undiscovered binding patterns.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
11/For example, while previous literature reported that JUND is "uniformly inhibited by 5mC", our analysis suggests that a minority of JUND motifs have a slight preference for 5mC. pic.twitter.com/R3SmAztSmz
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
12/FEZF2 appears to have two completely different motifs, one which prefers modified cytosine, and another which prefers unmodified cytosine. pic.twitter.com/HYh6bieyJn
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
13/For OCT4, our expanded alphabet analysis revealed a potential preference for 5-hydroxymethylcytosine. Unlike some of the other preferences described above, we could not find this described in the literature. pic.twitter.com/RmfVrj8MFn
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
14/Everything above is correlative—we were looking at existing ChIP and bisulfite data for the same cell types, but collected at different times. That doesn't prove that the binding site in ChIP actually has modified or unmodified cytosines.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
15/This observation was intriguing enough that we wanted to examine whether we could validate it directly, and we set out to do that before submitting the paper to a journal for the first time.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023
If only we had known that a global pandemic would be starting soon. -
16/To do this, @CharlesIshak @roxs88 in @decarvalho_lab and Santana Lardo in @HainerLab did CUT&RUN on OCT4 in mouse embryonic stem cells. The CUT&RUN generated DNA fragments from which we prepared three sequencing libraries: conventional, bisulfite-converted, and hmC-Seal-seq.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
17/This methodology gives us evidence of the epigenetic modification state of the DNA that is actually bound to OCT4. And, as it turns out, OCT4 motifs are indeed preferentially bound when the sequence is hydroxymethylated! pic.twitter.com/TjrRHLpir2
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
18/The Cytomod software that takes modification data and creates a modified expanded-alphabet genome sequence is on Zenodo and GitHub: https://t.co/JmJWu9cWh2
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
19/The MEME::Alphabet Perl module can be used to add expanded alphabets to non-MEME software. It is on CPAN. https://t.co/Ma4Xwmy5Mg
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
20/Other analysis scripts are on @github and @ZENODO_ORG: https://t.co/mQrTu5f6BU
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
21/The full set of scores generated in our analysis are available @ZENODO_ORG: https://t.co/s3QgbjvJSr
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 17, 2023 -
24/Conflicts of interest: an author is an inventors on patent applications for technologies that measure and analyze DNA modifications, filed by @CEGX_news, and hold stock options in @CEGX_news.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 19, 2023 -
25/Authors, including myself, are inventors on patent applications related to cell-free DNA methylation analysis technology, licensed to @adela_bio. Authors, not including myself, serve in leadership roles at and own equity in @adela_bio.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 19, 2023 -
26/Also, special thanks to @NewPI_Slack for introducing me to @HainerLab! Without her help, the experimental part wouldn't have been possible.
— Michael Hoffman @michaelhoffman@mastodon.social (@michaelhoffman) March 19, 2023
