UK Bat Coronaviruses Preprint Update
An update to a preprint on UK bat coronaviruses has revealed some unexpected and surprising results. Four species of coronaviruses were recovered from a small sample of 48 droppings from 16 of the 17 bat species breeding in the UK.

Prof Francois Balloux
Director @UGI_at_UCL. Interest in Infectious disease epidemiology, pathogen genomics and global health Mastodon account: @FBalloux@genomic.social
-
We've updated our preprint on UK bat coronaviruses. A lot of additional work went into it and there are new results, some pretty unexpected and surprising.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
1/https://t.co/JxDEUfHBOn -
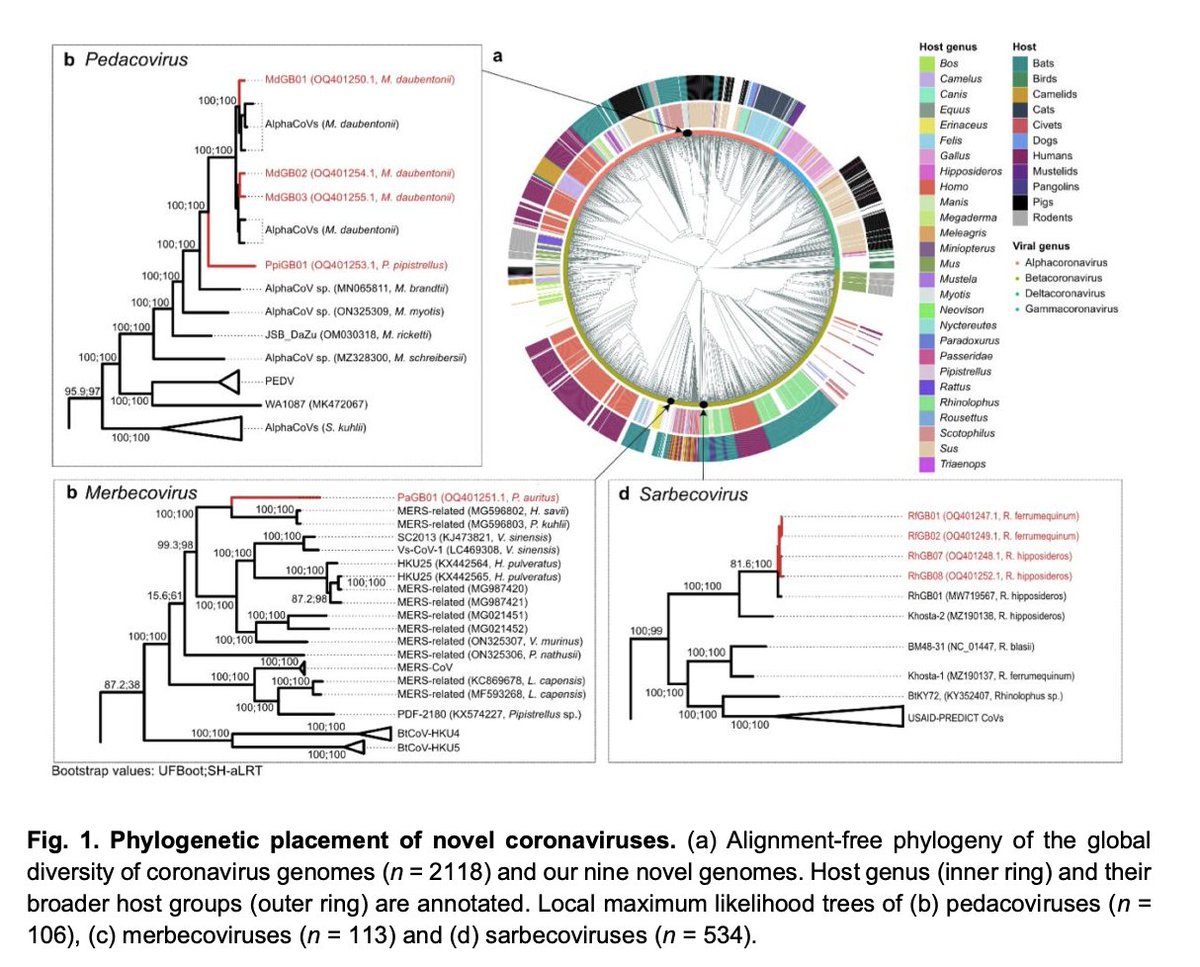
What didn't change. We recovered complete high-quality genomes for 4 species of coronaviruses (2 novel) in a small sample of 48 droppings from 16 of the 17 bat species breeding in the UK. There are likely more coronaviruses in circulation in UK bats.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
2/ pic.twitter.com/d6fIRmdvuU -
None of the coronaviruses, 2 Alphas, 1 MERS-like, and 1 sarbecorvirus (loosely related to SARS-CoV-2) can readily infect human cells. But ...
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
3/ pic.twitter.com/SyF7K2gVaK -
One bat sarbecovirus we tested could bind and use human ACE2 when overexpressed (ACE2 is also the cell receptor used by SARS-CoV-2). Thus, the UK bat sarbecovirus might have some 'zoonotic potential', but it would require further molecular adaptations to infect humans.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
4/ -
The spike protein of the UK bat sarbecovirus is structurally very similar to SARS-CoV-2, but the residues that bind to ACE2 are poorly conserved. Interestingly, the UK bat sarbecoviruses are only nucleotide mutation away from a canonical furin site sequence.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
5/ pic.twitter.com/AbPB9nyjWu -
Though, when we mutated the UK bat spike sequence to a canonical furin site, it wasn't functional. This likely stems from the lack of a 'spaghetti loop' that allows SARS-CoV-2 to expose its furin cleavage site.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
6/ -
Another surprising result is that the UK bat sarbecoviruses don't seem to use ACE2 in Rhinolophus bats, their natural host reservoir. We have at this stage no idea what cell receptor they may use to infect their natural host.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
7/ -
Before anyone freaks out, please let me reassure you no bat got hurt during the work. We used bat faecal samples (droppings). Also, none of the experiments were dangerous in any way. We didn't use any live virus. All lab experiments were done with totally safe 'pseudoviruses'.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
8/ -
This was a massively collaborative and interdisciplinary effort. I can't thank everyone here, and highlighting anyone's contribution in particular may feel unfair to other members of the team. Thus, please have a look at the author list and acknowledgment section.
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
9/ -
In tweet 5, this was meant to read:
— Prof Francois Balloux (@BallouxFrancois) March 22, 2023
" (...) only *one* nucleotide mutation away from a canonical furin cleavage site.".
5b/
