Mapping Mouse Prenatal Development with Single Cell Resolution
A collaboration between researchers from the Chengxiang Qiu lab and the Jackson Lab has yielded a preprint on mapping mouse prenatal development with single cell resolution. This research provides insight into the stages of development from single cell zygote to free-living pup.

Jay Shendure
Genomics technology developer. Developmental biology adult learner. Professor at @uwgenome @HHMINEWS @BrotmanBaty. He/him.
-
Excited to share our lab's latest preprint, led by @CXchengxiangQIU, @bethkarenmartin & Ian Welsh of @jacksonlab.
— Jay Shendure (@JShendure) April 6, 2023
We set out to build a single cell roadmap for all of mouse prenatal development, from single cell zygote to free-living pup.
Preprint: https://t.co/GDS65Qf58X 1/n pic.twitter.com/LtNcaZ1l5p -
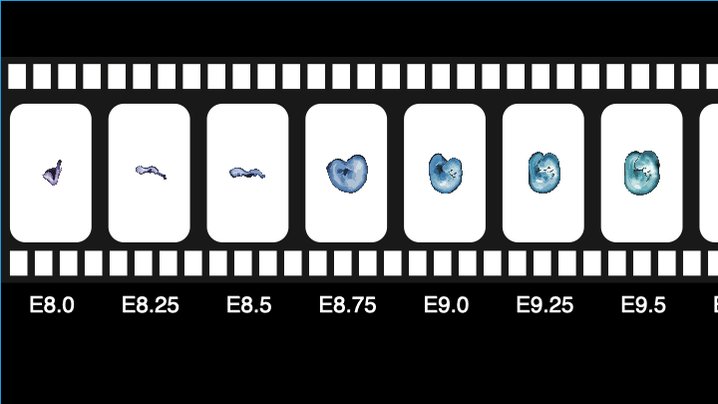
This was a collaboration with @jacksonlab, where Ian Welsh precisely staged 523 embryos spanning E8 to P0, identifying 74 that would give us 2 hour temporal resolution during somitogenesis (0-34 somite counts) and 6 hour resolution through to birth. 2/n pic.twitter.com/AYXflEiK9A
— Jay Shendure (@JShendure) April 6, 2023 -
The embryos were shipped to the remarkable @bethkarenmartin in Seattle, who applied her optimized sci-RNA-seq3 protocol (original by the brilliant @junyue_cao @RockefellerInst) to profile 12.4 million single nuclei (median 2545 UMIs per nucleus). 3/n pic.twitter.com/ovy66ExDV4
— Jay Shendure (@JShendure) April 6, 2023 -
After identifying & annotating cell types, @CXchengxiangQIU performed deeper dives into ontogenesis of the posterior gastrula, kidney, mesenchyme, retina, early neurons & other systems. I can't do these justice here; please see preprint for details (https://t.co/GDS65Qf58X) 5/n pic.twitter.com/cjkkB2BvNA
— Jay Shendure (@JShendure) April 6, 2023 -
How to summarize the complexity of development? @CXchengxiangQIU developed a simple heuristic for linking cell types based on excessive sharing of temporally coincident, mutual nearest neighbors in PCA space. 6/n pic.twitter.com/wmD9dqUK3I
— Jay Shendure (@JShendure) April 6, 2023 -
The complexity of mammalian development is such that this required some manual curation, but we broke down the problem by wrangling each of 12 subsystems separately, e.g. hematopoiesis. 7/n pic.twitter.com/aUeBJXVovi
— Jay Shendure (@JShendure) April 6, 2023 -
Building on what had worked for us before (https://t.co/bwcdN2KGfb), we also applied this approach to 4 other sc-RNA-seq datasets that collectively traversed development from zygote to gastrula, and put these all together. 8/n pic.twitter.com/sdyLJwdm5X
— Jay Shendure (@JShendure) April 6, 2023 -
The result is a cell type tree that traverses mouse development from single cell zygote to free-living pup. 9/n pic.twitter.com/nSxzChViVU
— Jay Shendure (@JShendure) April 6, 2023 -
What surprised us the most in these data? We oddly observed that in some cell types, there were MASSIVE differences between E18.75 and P0 mice. 10/n pic.twitter.com/nJ5Hqld3ay
— Jay Shendure (@JShendure) April 6, 2023 -
Technical or analytical artifact, perhaps? Validation experiments confirmed the finding, and narrowed its initiation to the hour immediately following birth. 11/n pic.twitter.com/ntSHZOSmH5
— Jay Shendure (@JShendure) April 6, 2023 -
What's going on here? On brief reflection, it's immediately apparent that one of these things is not like the other. (credit to R. Bejema for illustrations) 12/n pic.twitter.com/U1ToBXdd0w
— Jay Shendure (@JShendure) April 6, 2023 -
In particular, the transition from the uterus to the extrauterine world is a rather large change in circumstances for any mammal. 13/n pic.twitter.com/0UFgTgyfsH
— Jay Shendure (@JShendure) April 6, 2023 -
We speculate that these rapid, cell type-restricted changes are poised physiological adapations that are triggered by birth, e.g. gluoconeogenesis in the liver (cut off from mom), thermogenesis by fat (the real world is cold), etc. 14/n pic.twitter.com/04jyML7vhv
— Jay Shendure (@JShendure) April 6, 2023 -
Why did we do this, and where might we go next? Again please see the preprint but also a somewhat philosophical perspective that @sdomcke and I wrote that tries to capture the broader context for this line of work. 15/nhttps://t.co/NLAPdHcwFl
— Jay Shendure (@JShendure) April 6, 2023 -
Many people to thank, but in particular this work was largely executed by three people: @CXchengxiangQIU (all of the analysis!), @bethkarenmartin (vast majority of the data generation!) and Ian Welsh at JAX (all of the embryo staging!). 16/n
— Jay Shendure (@JShendure) April 6, 2023 -
Building on longstanding collaborations with @coletrapnell, experimental methods from @junyue_cao, the vision of @malte_spielmann, and a wonderful collaboration w/ Steve Murray's lab @jacksonlab. 17/n
— Jay Shendure (@JShendure) April 6, 2023 -
Copious guidance from @schierlab @Nobu_Hamazaki @CeciliaMoens @thabangh @SRsrivatsan @evaknichols (& others not on Twitter or whose handles I missed). And a huge thank you to @BrotmanBaty @AllenInstitute @HHMINEWS @jacksonlab @NIHFunding for funding the work. 18/n
— Jay Shendure (@JShendure) April 6, 2023 -
I'll be talking about this work tomorrow at @satijalab's Single Cell Genomics Day. We welcome any feedback on the preprint, and thank you for reading this far! 19/nhttps://t.co/VBCYV7ok8O
— Jay Shendure (@JShendure) April 6, 2023
